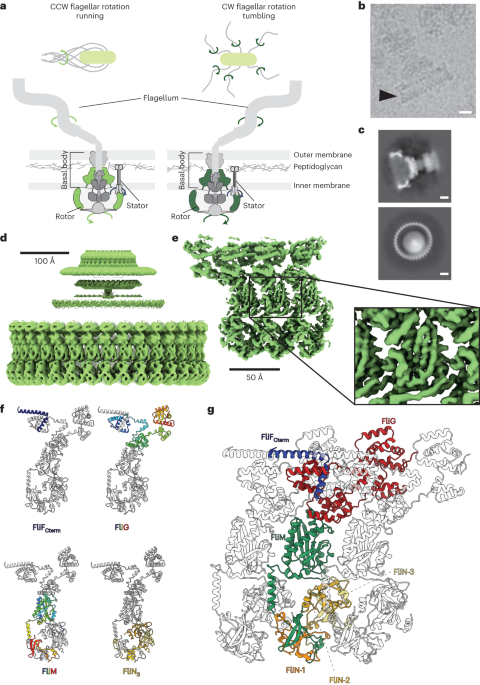
Berg, H. C. Bacterial behaviour. Nature 254, 389–392 (1975).
Berg, H. C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 (2003).
Nakamura, S. & Minamino, T. Flagella-driven motility of bacteria. Biomolecules https://doi.org/10.3390/biom9070279 (2019).
Carroll, B. L. & Liu, J. Structural conservation and adaptation of the bacterial flagella motor. Biomolecules https://doi.org/10.3390/biom10111492 (2020).
Minamino, T. & Imada, K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 23, 267–274 (2015).
Johnson, S. et al. Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation. Nat. Microbiol. 5, 966–975 (2020).
Berg, H. C. Torque generation by the flagellar rotary motor. Biophys. J. 68, 163S–166S (1995).
Minamino, T., Morimoto, Y. V., Hara, N., Aldridge, P. D. & Namba, K. The bacterial flagellar type III export gate complex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoS Pathog. 12, e1005495 (2016).
Manson, M. D., Tedesco, P., Berg, H. C., Harold, F. M. & Van der Drift, C. A protonmotive force drives bacterial flagella. Proc. Natl Acad. Sci. USA 74, 3060–3064 (1977).
Wadhams, G. H. & Armitage, J. P. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037 (2004).
Deme, J. C. et al. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat. Microbiol 5, 1553–1564 (2020).
Santiveri, M. et al. Structure and function of stator units of the bacterial flagellar motor. Cell 183, 244–257.e16 (2020).
Leake, M. C. et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358 (2006).
Lele, P. P., Hosu, B. G. & Berg, H. C. Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl Acad. Sci. USA 110, 11839–11844 (2013).
Nord, A. L. et al. Catch bond drives stator mechanosensitivity in the bacterial flagellar motor. Proc. Natl Acad. Sci. USA 114, 12952–12957 (2017).
Reid, S. W. et al. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl Acad. Sci. USA 103, 8066–8071 (2006).
Wadhwa, N., Sassi, A., Berg, H. C. & Tu, Y. A multi-state dynamic process confers mechano-adaptation to a biological nanomachine. Nat. Commun. 13, 5327 (2022).
Francis, N. R., Sosinsky, G. E., Thomas, D. & DeRosier, D. J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235, 1261–1270 (1994).
Thomas, D. R., Morgan, D. G. & DeRosier, D. J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl Acad. Sci. USA 96, 10134–10139 (1999).
Thomas, D. R., Francis, N. R., Xu, C. & DeRosier, D. J. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188, 7039–7048 (2006).
Carroll, B. L. et al. The flagellar motor of Vibrio alginolyticus undergoes major structural remodeling during rotational switching. eLife https://doi.org/10.7554/eLife.61446 (2020).
Chang, Y. et al. Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 27, 1041–1047 (2020).
Minamino, T., Kinoshita, M. & Namba, K. Directional switching mechanism of the bacterial flagellar motor. Comput. Struct. Biotechnol. J. 17, 1075–1081 (2019).
Chang, Y., Carroll, B. L. & Liu, J. Structural basis of bacterial flagellar motor rotation and switching. Trends Microbiol. 29, 1024–1033 (2021).
Afanzar, O. et al. The switching mechanism of the bacterial rotary motor combines tight regulation with inherent flexibility. EMBO J. 40, e104683 (2021).
Yuan, J. & Berg, H. C. Ultrasensitivity of an adaptive bacterial motor. J. Mol. Biol. 425, 1760–1764 (2013).
Fukuoka, H., Sagawa, T., Inoue, Y., Takahashi, H. & Ishijima, A. Direct imaging of intracellular signaling components that regulate bacterial chemotaxis. Sci. Signal 7, ra32 (2014).
Young, H. S., Dang, H., Lai, Y., DeRosier, D. J. & Khan, S. Variable symmetry in Salmonella Typhimurium flagellar motors. Biophys. J. 84, 571–577 (2003).
Baker, M. A. et al. Domain-swap polymerization drives the self-assembly of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 23, 197–203 (2016).
McDowell, M. A. et al. Characterisation of Shigella Spa33 and Thermotoga FliM/N reveals a new model for C-ring assembly in T3SS. Mol. Microbiol. 99, 749–766 (2016).
Paul, K., Brunstetter, D., Titen, S. & Blair, D. F. A molecular mechanism of direction switching in the flagellar motor of Escherichia coli. Proc. Natl Acad. Sci. USA 108, 17171–17176 (2011).
Kinoshita, M. et al. Insight into adaptive remodeling of the rotor ring complex of the bacterial flagellar motor. Biochem. Biophys. Res. Commun. 496, 12–17 (2018).
Irikura, V. M., Kihara, M., Yamaguchi, S., Sockett, H. & Macnab, R. M. Salmonella Typhimurium FliG and FliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175, 802–810 (1993).
Welch, M., Oosawa, K., Aizawa, S. & Eisenbach, M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl Acad. Sci. USA 90, 8787–8791 (1993).
Lee, S. Y. et al. Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 8, 52–56 (2001).
Dyer, C. M., Vartanian, A. S., Zhou, H. & Dahlquist, F. W. A molecular mechanism of bacterial flagellar motor switching. J. Mol. Biol. 388, 71–84 (2009).
Antani, J. D. et al. Mechanosensitive recruitment of stator units promotes binding of the response regulator CheY-P to the flagellar motor. Nat. Commun. 12, 5442 (2021).
Bai, F. et al. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science 327, 685–689 (2010).
Johnson, S. et al. Molecular structure of the intact bacterial flagellar basal body. Nat. Microbiol. 6, 712–721 (2021).
Palovcak, E. et al. A simple and robust procedure for preparing graphene-oxide cryo-EM grids. J. Struct. Biol. 204, 80–84 (2018).
Caesar, J. et al. SIMPLE 3.0. Stream single-particle cryo-EM analysis in real time. J. Struct. Biol. X 4, 100040 (2020).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife https://doi.org/10.7554/eLife.42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Asarnow, D., Palovcak, E. & Cheng, Y. UCSF pyem v0.5. Zenodo https://doi.org/10.5281/zenodo.3576630 (2019).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).

Dr. Thomas Hughes is a UK-based scientist and science communicator who makes complex topics accessible to readers. His articles explore breakthroughs in various scientific disciplines, from space exploration to cutting-edge research.